What DNA Can Teach Us
We know it’s impolite to ask someone’s age – but for marine mammals it might be a critical question for conservation.
Research shows that as marine and terrestrial mammals age, DNA begins to show signs of degradation. Over time the physiological mechanisms in place to protect and preserve DNA from internal or external factors grow less effective. This includes the effects that environmental pollutants have on marine mammals. By studying the effects of aging on DNA at specific places amongst species, scientists have recently been able to develop DNAm epigenetic clocks that are strongly correlated with chronological age in humans and several mammalian species.
Developing epigenetic clocks from known age animals, like those living in zoos, aquariums or known age animals from the wild, scientists can then apply the results to accurately determine the age distribution among wild populations.
Dr. Todd Robeck from SeaWorld, in collaboration with geneticist and biostatisticians from UCLA, has been leading the application of this technology to toothed whales and dolphins. Dr. Robeck is currently the Vice President of Conservation Research and Animal Health at SeaWorld Parks & Entertainment. Recently Dr. Robeck sat down with Grey Stafford of the Zoo Logic podcast to discuss the recent collaborative work SeaWorld is undertaking across several species of odontocetes. This type of marine research promises to provide scientists and government agencies a powerful marine conservation tool.
Zoo Logic is brought to you by Animal Care Software. Take a test drive for your facility by visiting animalcaresoftware.com, using the coupon code, ZooLogic. Zoo Logic is also brought to you by kongzoo.com. Get an additional 10% off your well below retail pricing on your next order of enrichment items at kongzoo.com, using the coupon code, ZooLogic10.
Coming up on the podcast today, we're talking about epigenetic clocks. It's a science that's been around for a few decades with regard to human beings. But lately it's being applied to mammalian species. Today's guest is going to talk about this clock that's been developed for odontocetes, or toothed cetaceans, and what are the conservation and wildlife biology implications of having such a tool. And of course we've got an all new That Sounds Wild.
We are very fortunate today to have with us a veterinary expert, particularly in the area of reproductive medicine. His name is Dr Todd Robeck, and he currently serves as the Vice President of Conservation Research and Animal Health for our good friends at SeaWorld and Busch Gardens.
Dr Todd, welcome to Zoo Logic.
But anyway, these changes that occur actually on the DNA strands themselves and they occur somewhat random. But the location that the changes, or what essentially it's a methylation that is occurring, transferring of a methyl group, typically from an adenine to a cytosine.
And when that happens, depending on where it occurs in relationship to genes, promoter regions on the genes, or different areas on the genes that either turns the genetic function on to produce proteins or turns it off, it can then change the expression of these genes which then go on to RNA or messenger RNA and then produce proteins that regulate essentially your body function.
So as you age -- and this is well known fact of aging and has been over some time, that as you age, your DNA is damaged either from just internal normal physiologic processes or from external processes that can cause damage to your DNA, also pollutants, environmental pollutants, toxicants, stress, all kinds of physiologic pressures that occur externally and internally.
So the trick is to then identify what changes that have occurred are important for aging or that relate to aging. Because remember, these changes occur throughout the DNA. So then you have to identify exactly where these changes have occurred, and if they actually relate to the age, the biologic or chronological age of that animal in this case.
Steve Horvath at UCLA has developed an array that can identify approximately or close to 38,000 different cytosine locations on your DNA, and determine their methylation state.
This is a collaboration with his group out of UCLA. If you look at the paper, you see a number of their coauthors, and they are the geneticists who are at the heart of the study.
And working with them and their technology, that we've put together this clock that's specific for odontocetes in this case, where we looked at nine different species of odontocetes. And then from that we were able to combine their methylation patterns, and in the fact that we knew their chronological age, were able to come up with a clock or a curve that then we can take an unknown animal and determine their methylation rate on these specific sites, also known as CPGs. And by determining the amount that of methylation that's occurred, then we can put it onto this, quote, "clock curve" and determine the age of that particular animal.
Is it good? I don't think generally changes to DNA is typically thought of as good. There are therapeutics that are being looked at to reduce the amount of methylation that is occurring.
You got to remember, I'm not a geneticist and I don't study methylation patterns in the organism as my expertise. So I can't answer if there is any good methylation going on. But my feeling on what I have looked at or read over the last couple of years as I've delved into this research is that it's not good or bad. And where it happens really determines if causes a detrimental effect on the organism.
So, you have methylation that naturally occurs as you live, as I said before. Food has to be oxidized, and that has to be turned into energy, and there's a whole -- cytochrome p450 pathway using oxygen, why we have to rely on oxygen, produces ATP. And in that process of producing energy, which is also known as ATP, you get radical oxygen species that can then actually damage the adjacent DNA. Most of that occurs in the mitochondria.
So in the cell itself, you can get -- there's different mechanisms to deal with the oxygenated radical oxygen species that could cause damage.
And as you get older, some of these systems that are supposed to claim these free oxygen species, they become less effective because of damage to them themselves -- and all kinds of hypotheses. But ultimately, methylation is a normal process whereby you live.
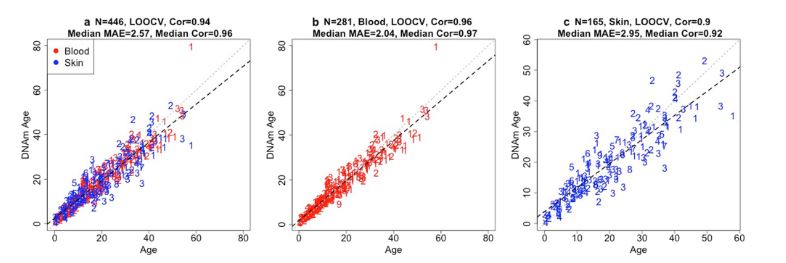
Now, one of the things about this study that's interesting is that we're categorizing what could be considered normal methylation rates with age. So that's why we're relating, we're picking out the changes that have occurred that then can be related, directly correlated, and correlation results are way over 90% for most of the species, and for the clocks themselves they are approaching 95%, especially in the blood.
These changes are relatable in every animal, at every age, just normally. So that's how we can determine their age.
There is something called accelerated aging, and that's more your biologic age. So that's something that we may have all heard -- what is your biologic age? And does that relate, or is that consistent with your chronological age? In other words -- are you a 60 year old that that has the biology of a 40 year old? Or are you a 60 year old that has a biology of an 80 year old?
And of course that can be affected by not only your own ability to handle oxidative stress in your life, but also your ability, what you've been exposed to externally, environmentally.
That ultimately -- this research has two fundamental purposes. One is that with this clock, we can say, get a blood sample or a skin sample from animals in the wild, or in a semi reserve situation, and determine their age very accurately, within 4 to 5 years of age. And especially with long lived species, the odontocetes that the toothed whales and dolphins are, that's very accurate. For a 50 year old animal, we can tell if he's 45, or up to 54 or 55, within a range there, for say, a skin sample.
And what that means is that then we know what age group... We know, if that animal is a female, is she likely to be fertile? Is she likely to be able to contribute to that population in the future? Or is she reaching the end of a reproductive life, or experiencing reproductive senescence? And and also we can predict, well, how long is that animal going to actually contribute to the functioning of that population in the situation they're in? In other words, are they going to contribute to learning of the adolescents? Are they going to help with foraging? Are they going to be feeding? So you're going to be at, this sort of a mass effect of the biology of that population is very important.
So all that information is critical to predict the current health of that species or that population of a species in the area that they are residing. So, are they at risk of becoming extinct because they are a very old population? Or do they have a lot of young animals with the potential to reproduce and continue that population on into the future? So, those are some a fairly fundamental population biology questions that now can be answered without having to pull an animal out of the water, get their age by sectioning they're tooth, which is typically how these these procedures were done in the past. So it's a very useful tool from that standpoint.
The next important point is that once we have an understanding of, and are able to detect normal chronologic age, we then can, say, look at a population of animals and try and determine if there is accelerated age happening? Are they being exposed to environmental issues that may be having a detrimental effect on their health and their biology, and thus are they older than they're supposed to be?
And we can do that a couple of ways. One way is, of course, if you have animals have known chronological age because you've been doing health assessments for 30, 40 years, so you have a large percentage of the population that have been identified at birth or as juvenile, so you have an accurate estimation of those animals -- then you can compare that estimated methylation age, which would be the estimated chronological age, versus real time age, and then if...
Of course, there's going to be individual variation, but if you see a trend within that population of animals... You got to remember, for this type of thing to work, it's more on a population level than an individual animal, because there will be individual variation. But if you see the population is out of sync with what is predicted, then you can say with extreme degree of confidence that something is happening to that population that is affecting, that is accelerating their age.
And then of course that's something is then whatever situation -- lack of food, high concentration of environmental pollutants or oxidants that are occurring in that area, all kinds of things, that then kind of fit with the location that's occurring.
The other thing you can do is, if you don't know their age, say you can get a sample of all the animals now, and then 5 or 10 years later, get another sample, and then you have a defined period of time whereby you can predict, well, they should be 10 years older. But what if they're 15 years older as a population, then you know, once again, oh, this population is under some kind of stress.
Then of course you have to go back and look into the environment where these animals are, their habitat and what they're experiencing, to understand potentially the link of significance.
You can also look at levels of pollutants -- pollutants is a very broad sort of inaccurate terminology -- but some kind of exposure to some chemicals or something that's causing oxidative stress or stress in the organism. So you can then look and correlate their accelerated age to the concentration of these chemicals in their body.
So, they've... They're doing that with, say, alcoholics in humans. And this is where most of the stuff was originally done. There's papers on looking at predicting cancer, cardiovascular disease, different types of mortality based on how accelerated their age is. You can look at... You can actually predict the time of death based on their methylation, and their health.
So, if you look at the human literature, DNA methylation is just really beginning to take off over the last few years, or epigenetic changes due to DNA methylation, and correlating it with actual disease.
That's the future of this. And this study we have done is forming sort of the ground level basis from which interpretations can be made at a population level concerning their overall health.
What is it about these questions that interest you so much, given your background as a veterinarian treating individual animals? You're now working at the population level.
So yeah, I have worked with individual animals. But also I have a PhD in reproductive physiology. So I have a lot of background physiology. And I have a degree in marine biology.
I initially started in my field, my interest in marine organisms, on more of a population level. So I've always been interested in the health of the oceans and how it's all interrelated.
And cetaceans are top level trophic species, whereby they -- and you've seen this all the time and it's sort of almost passe -- but they are an indicator species of the health of the ocean.
So, two things. One -- just my interest in marine biology alone. I mean, I could end my answer right there. But from a standpoint of working with zoologic species, is also interested in how is the zoological setting affecting the biology of these animals and how does it compare to the wild animals?
And and my sort of working hypothesis is that, I mean, they're not necessarily better, they're just different. And these animals, I have believed, and I have been able to demonstrate most of the time, that they provide a sort of a warehouse of... Not only are they ambassador animals for people to be exposed to them in the first place, but they're also a group of animals that can help provide answers that can then directly affect and understand changes that occur in wild populations.
This study is no exception. This study cannot be done effectively without known age animals. So we've put together over 400. I mean, it's an amazing group of animals in nine different species, of known age or odontocetes, toothed whales, from different -- from beluga to killer whales, dolphins, pilot whales. We have just a tremendous sample set here, whereby we have known age or estimated, in other words they were neonates, juveniles, when when stranded, per se, or collected many, many years ago, since we haven't collected animals in so many years now -- so there were known-age animals or estimated age animals. Most majority of these used in this study, almost close to 80% were captive born.
So we have such a robust sample set, and then we have a nice sample set of known age killer whales from Norway also to include in this, from Norwegian Orca Survey. And Eve Jourdain has helped us out with that.
So we have a very robust sample set that is going to help of animals that have been followed and of known age that we can then use to directly apply to the health and well being of wild populations.
I don't think you can ask for a better opportunity as a marine biologist, as a reproductive physiologist, as a veterinary, whose concern, who's spent his whole life working with species in the wild and in zoo settings, a better opportunity to be involved with. So I'm actually very privileged to be sort of the lead with UCLA on this study.
And one of the things that I gleaned from your paper is -- the site selection on the DNA strands that are chosen and included as part of this Epigenetic Clock, they are conserved across species. In other words, these are common areas to all odontocetes, and perhaps even other species, other mammalian species at least.
Is that the case? The reason you're able to use 400 samples across several species is because you're selecting portions of DNA that are conserved among all those different species.
And his hypothesis when we first got involved with him was that he could develop a universal mammalian clock, which is now in in review right now, for, just as I said, any mammal species.
So our involvement at SeaWorld is not just SeaWorld but also Busch gardens. And we've provided samples, I'm not even sure the number, but it's approaching 200 different species. We've added to Steve's goal of developing a universal mammalian clock.
We've been very large significant contributors to this research. So this has been a collaboration across all our different zoos and aquariums to try and reach this goal. And part of it is the hypothesis that you can develop a universal clock. Therefore, the only way that's possible is if the aging process is conserved across species. And that's essentially what's coming out from this research.
There's another, I think, another paper that's currently in review that looks at using this information they have developed now, to be able to determine the maximum age that these different species can live.
So this is fairly exciting paper that's coming out and it's also under review, and it's by Steve's team, and so many different collaborators across the country and the world. It's quite a nice collaborative effort. It's epigenetic predictors of maximum lifespan and other life history traits and mammals.
So hopefully that's... It's already on Bioarchives as a pre print so people can look it up now and at least get a taste of it. But it's in formal review right now.
It's very interesting because, because of these conserve changes that occur across different species, we're then able to make a prediction. So if we look at supplemental file from that age, you're able to look at, he's got a correlation between maximum lifespan [predictor in samples. So they developed the predictor based on different CPG sites that have been methylated. And they've been able to fairly consistently identify maximum age.
So for some of the species whereby they have the most samples, they've actually put a graph out. So for instance, a dog, their predicted age is a little over 20. And they look at this prediction across the age range of the animal. So in other words, at a young age, they look at the methylation patterns and they can say, okay, well, at zero or 6 months of age, they predict the maximum lifespan of this animal is going to be 22. Well, they do it again at 10, at 10 years, say, well look, it's going to be 22. 15 years, well, look it's going to be 22. And in this case the dog works very well. They're able to predict very well the maximum estimated lifespan of a dog. I mean, there's individual animal differences in these predictions, but the data is very, very, very consistent and robust across this particular species. With a mouse, say you have about four years -- and this this graph is very interesting to me. With sheep, it is a little bit of a correlation with age, and that means they're not quite as accurate with this prediction. In other words, if you have a different age at one versus seven or eight, and if this change is happening across the age range, then they haven't identified all the different predictors of maximum age then. And there's some tweaking that still needs to be done, which of course they're working on all the time. So say a sheep, it's 25 at one age, and at 10 years of age, they're predicting it to be about 23. So there's a change, a little bit of a change is not exactly perfect, but pretty darn close. With a cow, you're getting a maximum age of a little over 30. With a killer whale, it's 70.
With a zebra, a zebra needs some work, maybe because of the sample size. Maybe just at 100 there. Goes from 40 to 33 with age. So clearly, there is some correlation with age on that with that species.
Beluga whale. They have at about 48 and that's very consistent across the age range. With a bottlenose dolphin, which of course, it has a little bit of correlation with age because it's about 55 at zero, and at about six years of age, it's down to about 50. So that's not a perfect correlation. There is a correlation there, just not a perfect predictor, but it's still pretty good. It doesn't change much.
So it's a really neat paper. And you got to remember, these are population level predictions. So you may have animals that live longer or shorter and it's just what the potential is for maximum age.
That's just one offshoot of this. You have the mammalian clock which Steve Horvath and his team is putting together that is a maximum predictor of age. And these are all offshoots of all this research.
And so why do we bother with doing this? If we have a universal clock for a mammalian clock, Why we bother with the odontocete clock? Well, it's simply because as you drill down into each species, your prediction accuracy will improve if you develop a clock within that individual species.
But if you don't have enough samples to increase your prediction accuracy, you want to increase your sample size.
So say, in odontocetes, we developed a clock with a number of different species. But for some of them we didn't have that many samples. So it would be impossible to develop an age predictor clock, say, on known age pilot whales, because we didn't -- I mean, if you had the samples, you could do it, but we didn't have that many. We had, I think 15 or so different samples of blood and skin samples.
But we can use that data within our study to develop a robust clock, that if we came into another pilot whale, we'd have fairly good confidence that we can predict the age of that animal.
But some of the other species we use, we have lots of samples, say, killer whales, beluga, bottlenose dolphins, pacific white sided dolphins. We have a number of samples there. And we could actually develop individual species clocks. But due to the constraints of the paper itself, and the size of it and the amount of information, it just becomes too unwieldy to publish a paper with all these different individual species clocks, and an odontocete clock.
So some of them, the species will come out with individual clocks. Some of them have already come out with the clock. There's a beluga paper that came out with a clock with Steve's lab, using just samples from wild animals.
And then we're in the process of publishing, or submitted a bottlenose dolphin clock to look at a bottlenose dolphin-specific skin and blood clock.
Is there a preferred type of tissue that this clock works best in? Like, if you had your druthers, what type of tissue would you use to go out and sample wild populations?
And blood samples are unbelievably accurate. They're 98% accuracy. I mean, it's just a very small deviation in years in terms of the range of age of the predicted age of animals.
So some of these questions about GLG counts... They're most controversial in old killer whales, and in beluga. Beluga across their whole age range, but also, they get difficult as they get older, because of the wear in the teeth. So, with beluga, especially, there's two different groups, felt like it was pretty widely accepted. It was two counts per year. So they had to growth that are groups per year. But this is the only species of odontocete that does that.
There are other groups -- why would the beluga have a different rate of deposition of cementum and dentin on their teeth as they grow than other odontocetes? It's a legitimate question. Why would they?
We don't really have the answer to that, or what we don't even know, actually, is if there is a different rate. So some groups have looked at captive animals and claim by their techniques... And there are also different skills involved in estimating the GLG counts. So some groups claim it's closer to two. And not exactly two. And there may be some deviation with age and growth rates. So there may be 2.2 at a younger age in 1.8 or 1.5 at an older age. I mean nobody knows.
But on average, about approximately two, versus one. And there are some groups who have come out with work that supports their idea that it's one.
Biologically speaking, which for me, is the most telling, is I look at estimates using two. And these animals are maturing reproductively at so much older than we know is the case from our animals at zoos.
So it, almost twice. So it's like, well, wait a minute, this fits right in here with two.
And also, then, we have animals that they're claiming, if you look at one per year, they reach upwards of 100 years of age.
And just, we know from a physiologic and biologic standpoint looking at our animals as they get older, you can see normal aging process in animals. And there's just no way it's off by a factor of two. And interesting.
So I think you have to put all this together. There's methodology issues, there is tooth wear issues, there's a lot of variations. So at this point I think using GLGs, especially in the older age groups, is not accurate.
But I don't have proof of that. I have som

